|
I am currently a PhD student at The Hong Kong University of Science and Technology, working under the supervision of Prof. Xiaomeng Li. Prior to this, I earned my Bachelor degree (with Honours) from The Australian National University, where I was fortunate to be advised by Senior Prof. Amanda Barnard and supervised by Dr. Amanda Parker. My research journey has been enriched by valuable experiences, including serving as a Research assistant at MIT Computational Connectomics Group led by Prof. Nir Shavit, where I had the privilege of collaborating with Prof. Hao Wang and Prof. Lu Mi. Additionally, I worked as a Research assistant at the National University of Singapore, supervised by Dr. Gang Guo. |

|
|
My research interest is to apply Interpretable Machine Learning to interdisciplinary fields.
|
|
"*" indicates equal contribution, "_" indicates equal advising. |
|
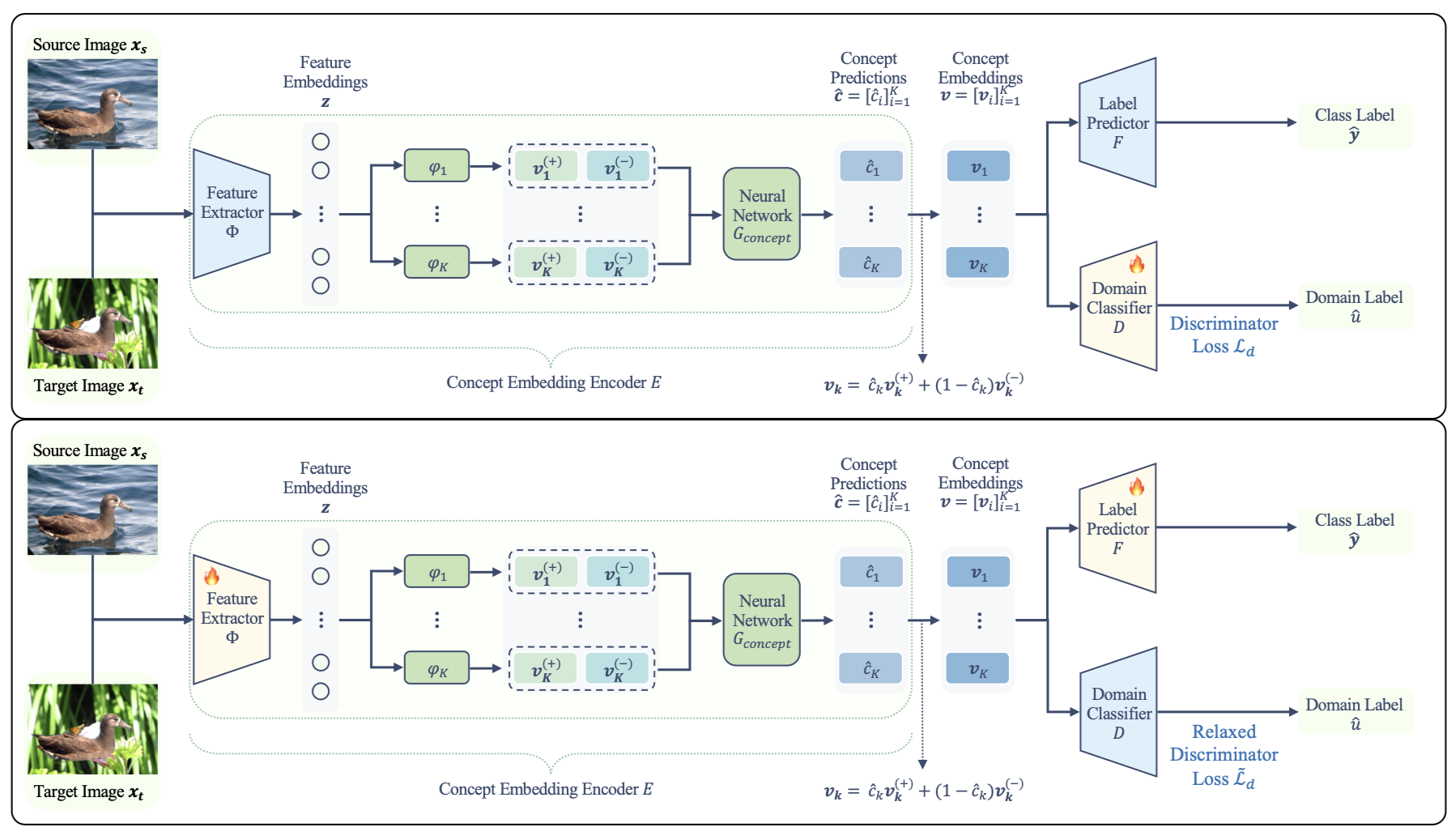
Xinyue Xu*, Yueying Hu*, Hui Tang, Yi Qin, Lu Mi, Hao Wang, Xiaomeng Li ICML, 2025 Paper / Code Interpretability We propose Concept-based Unsupervised Domain Adaptation, a framework that improves interpretability and robustness of Concept Bottleneck Models under domain shifts through adversarial training. |
|
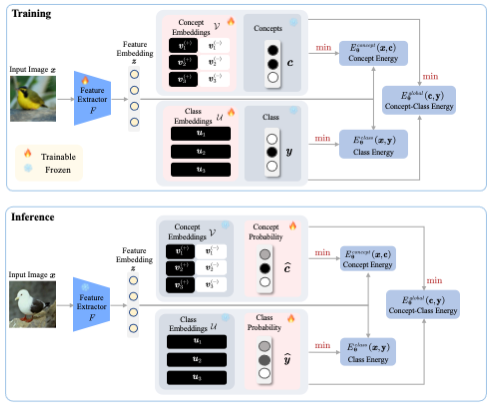
Xinyue Xu, Yi Qin, Lu Mi, Hao Wang, Xiaomeng Li ICLR, 2024 Paper / Code Interpretability We introduce Energy-Based Concept Bottleneck Models (ECBMs) as a unified framework for concept-based prediction, concept correction, and fine-grained interpretations based on conditional probabilities. |
|
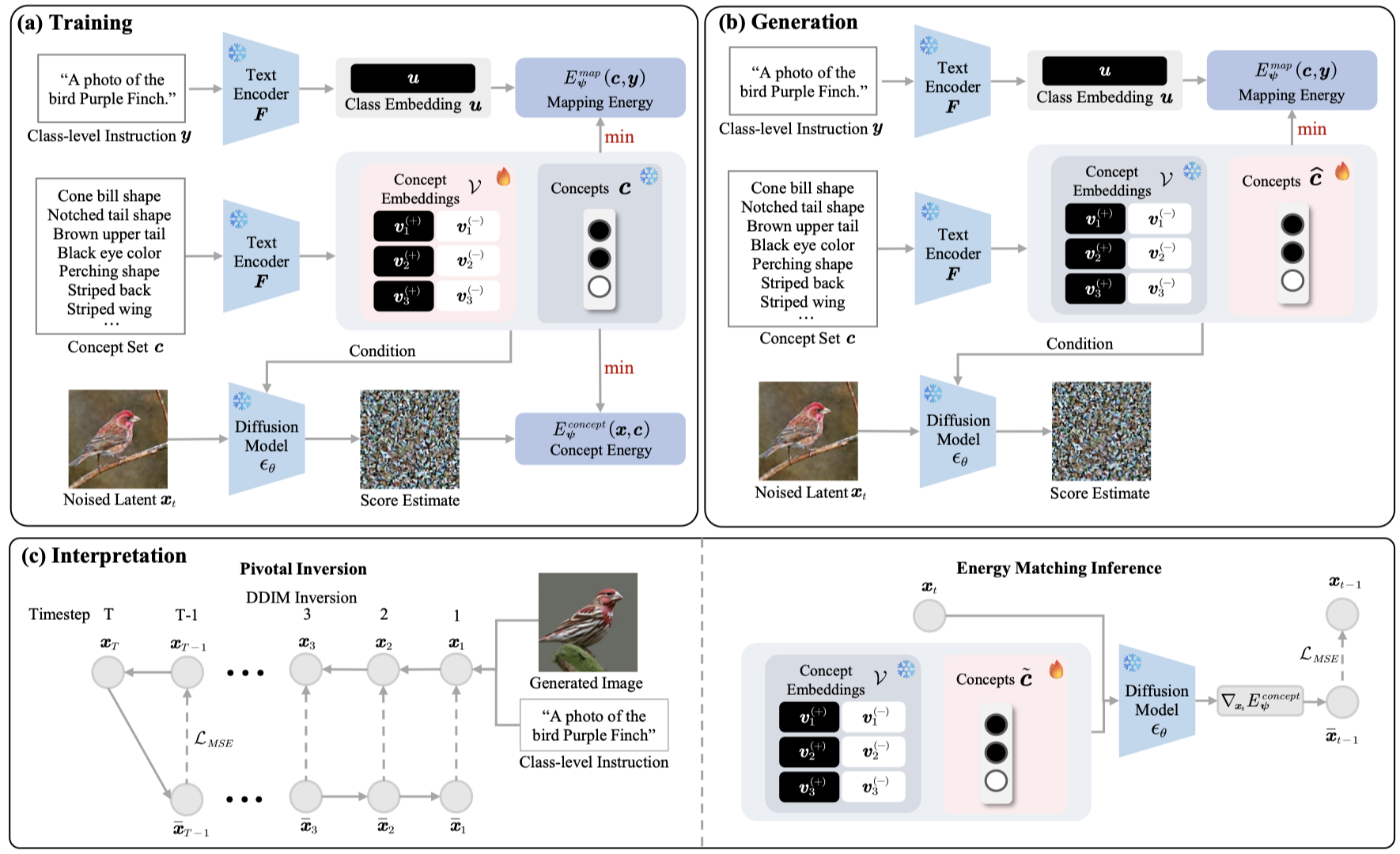
Yi Qin, Xinyue Xu, Hao Wang, Xiaomeng Li NeurIPS Safe Generative AI Workshop, 2024 Paper Interpretability We propose Energy-Based Conceptual Diffusion Models (ECDMs), a framework that unifies the concept-based generation, conditional interpretation, concept debugging, intervention, and imputation under the joint energy-based formulation. |
|
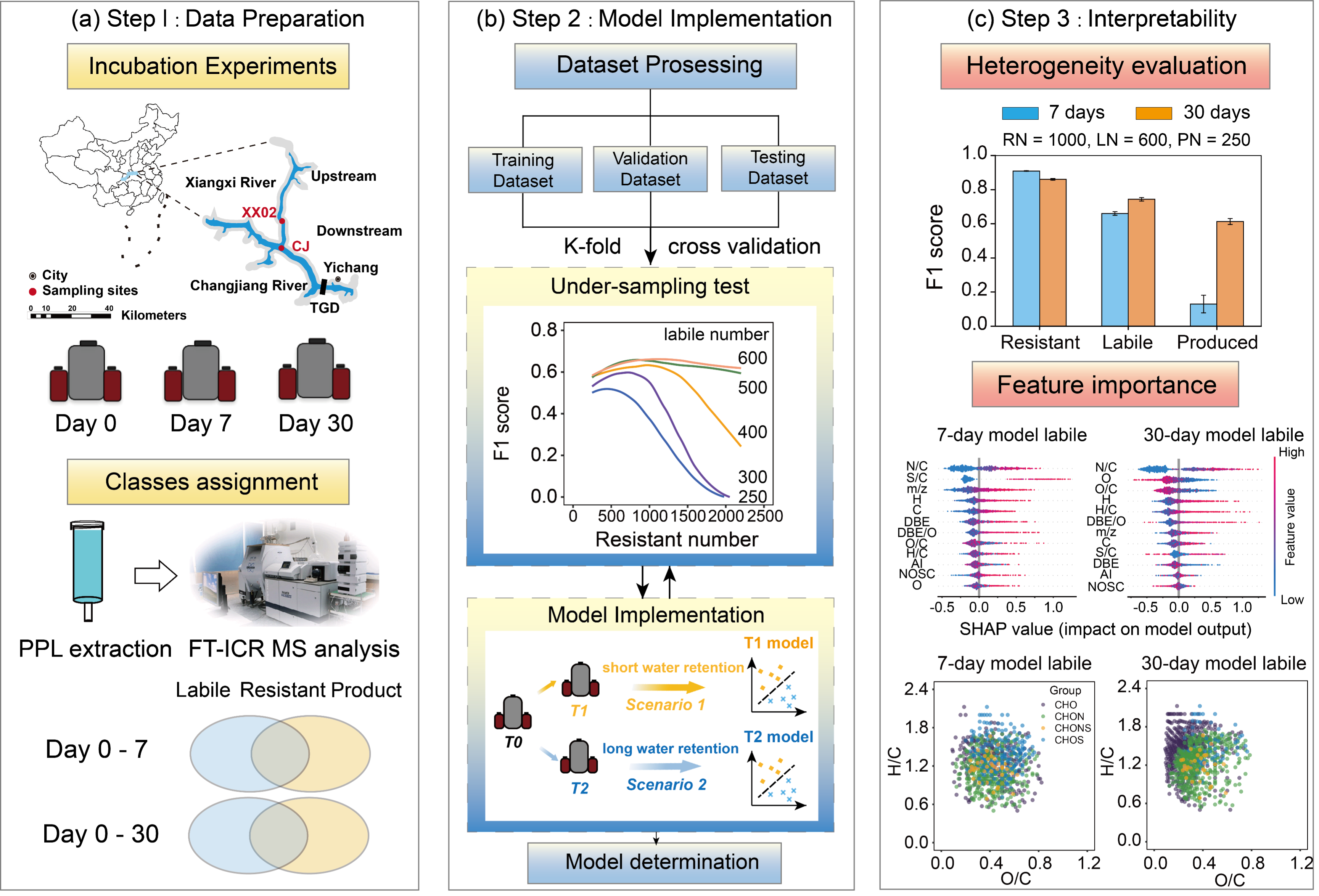
Chen Zhao, Kai Wang, Qianji Jiao, Xinyue Xu, Yuanbi Yi, Penghui Li, Julian Merder, Ding He Geophysical Research Letters, 2024 Paper AI4Science Interpretability Machine learning models were built to correlate the molecular composition and biological reactivity at the world's largest reservoir. Shorter incubations result in a broader range of molecules disappearing, with a greater contribution of stochasticity. Tuning the machine learning model contributes to yield additional interpretability beyond its well-recognized predictive power.
|
|
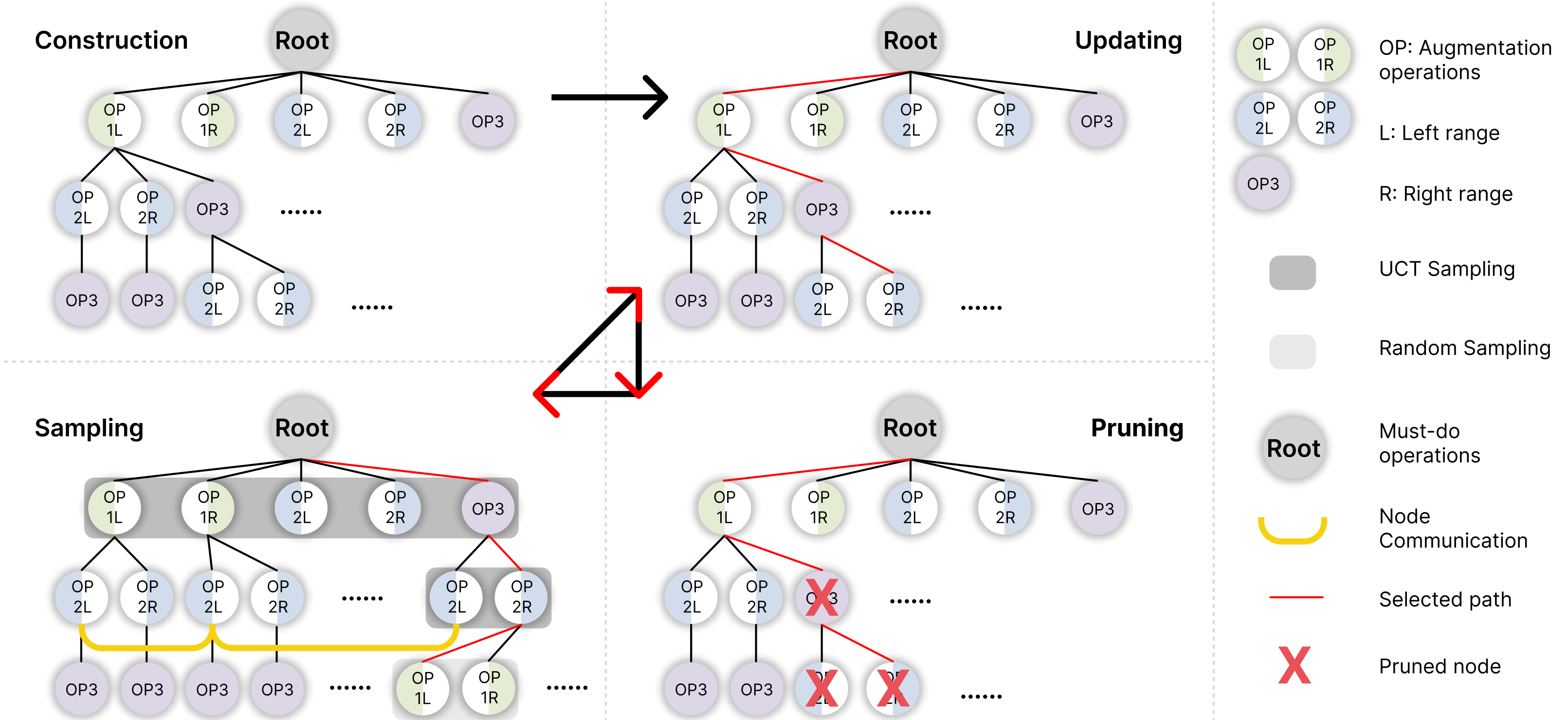
Xinyue Xu*, Yuhan Hsi*, Haonan Wang, Xiaomeng Li ICONIP, 2023 Paper / Code AI4Science Medical image data are often limited due to the expensive acquisition and annotation process. Hence, training a deep-learning model with only raw data can easily lead to overfitting. To this end, we present a novel method, called Dynamic Data Augmentation (DDAug), which is efficient and has negligible computation cost. |
|
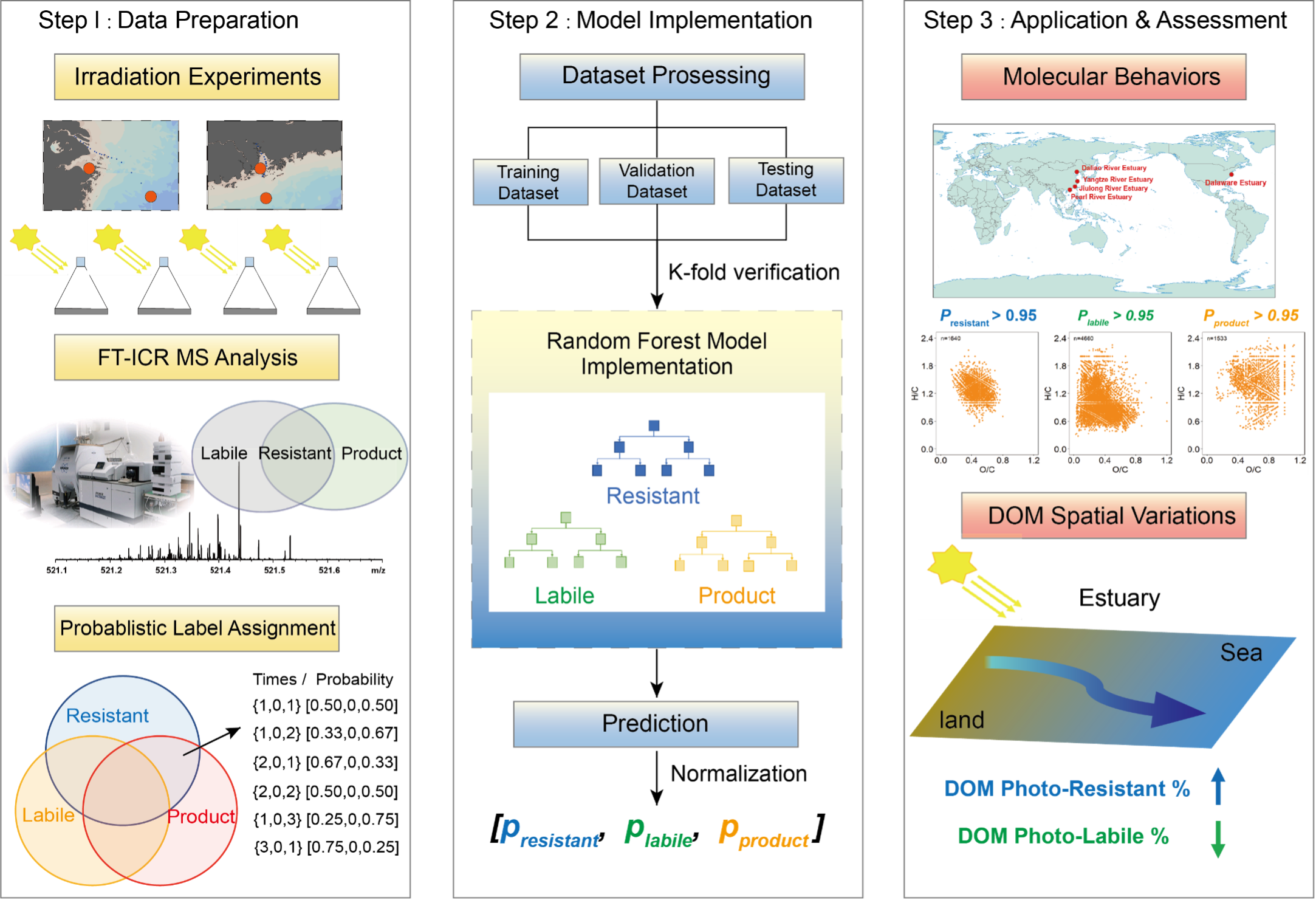
Chen Zhao*, Xinyue Xu*, Hongmei Chen, Fengwen Wang, Penghui Li, Chen He, Quan Shi, Yuanbi Yi, Xiaomeng Li, Siliang Li, Ding He Environmental Science & Technology (ES&T), 2023 Paper Improved Understanding of Photochemical Processing of Dissolved Organic Matter by Using Machine Learning Approaches (Conference Version) The 6th Xiamen Symposium on Marine Environmental Sciences, 2023 (Best Poster Award) AI4Science Interpretability Photochemical reactions are essential components altering dissolved organic matter (DOM) chemistry. We first used machine learning approaches to compatibly integrate existing irradiation experiments and provide novel insights into the estuarine DOM transformation processes. |
|
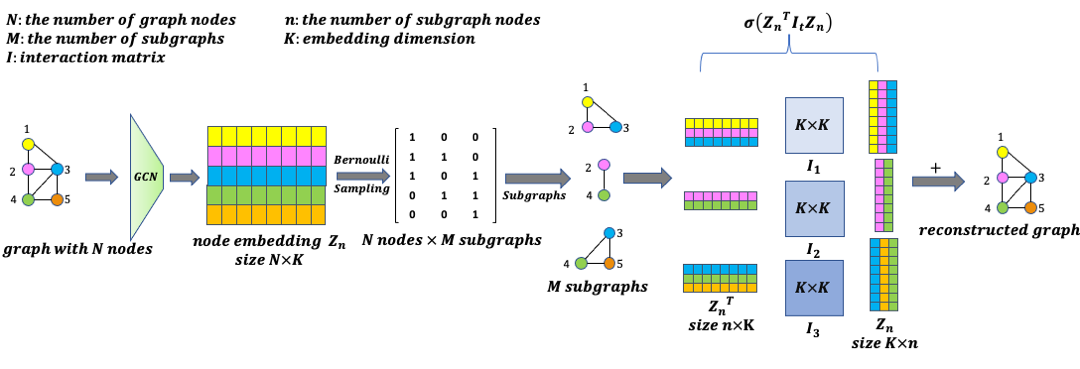
Zhongren Chen*, Xinyue Xu*, Shengyi Jiang, Hao Wang, Lu Mi KDD Deep Learning on Graphs, 2022 Paper AI4Science Small subgraphs (graphlets) are important features to describe fundamental units of a large network. Unfortunately due to the inherent complexity of this task, most of the existing methods are computationally intensive and inefficient. In this work, we propose GNNS, a novel representational learning framework that utilizes graph neural networks to sample subgraphs efficiently for estimating their frequency distribution. |
|
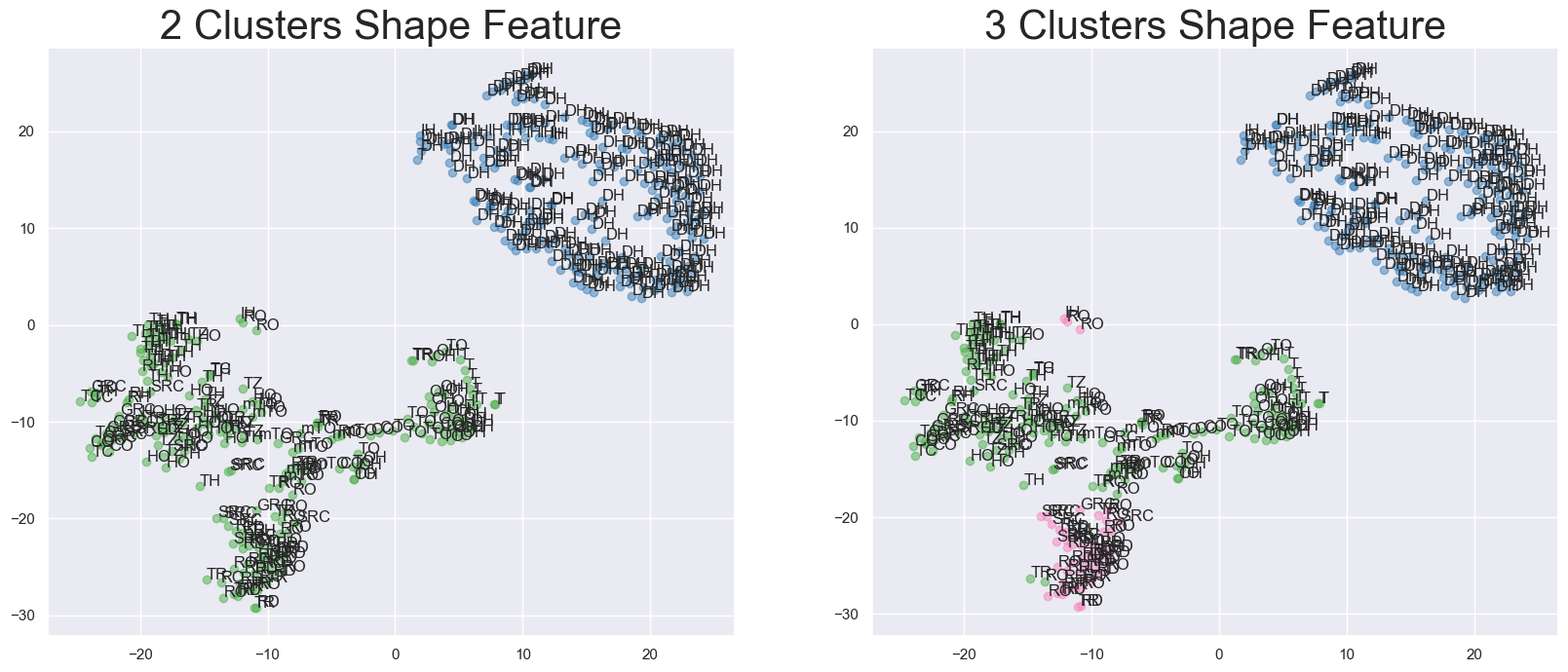
Xinyue Xu Honours Thesis, 2022 ANU Database. Code AI4Science This thesis develops an enhanced version based on the original Iterative Label Spreading Clustering which is specially designed for materials science, and aim to become faster and have fewer hyperparameters. |
|
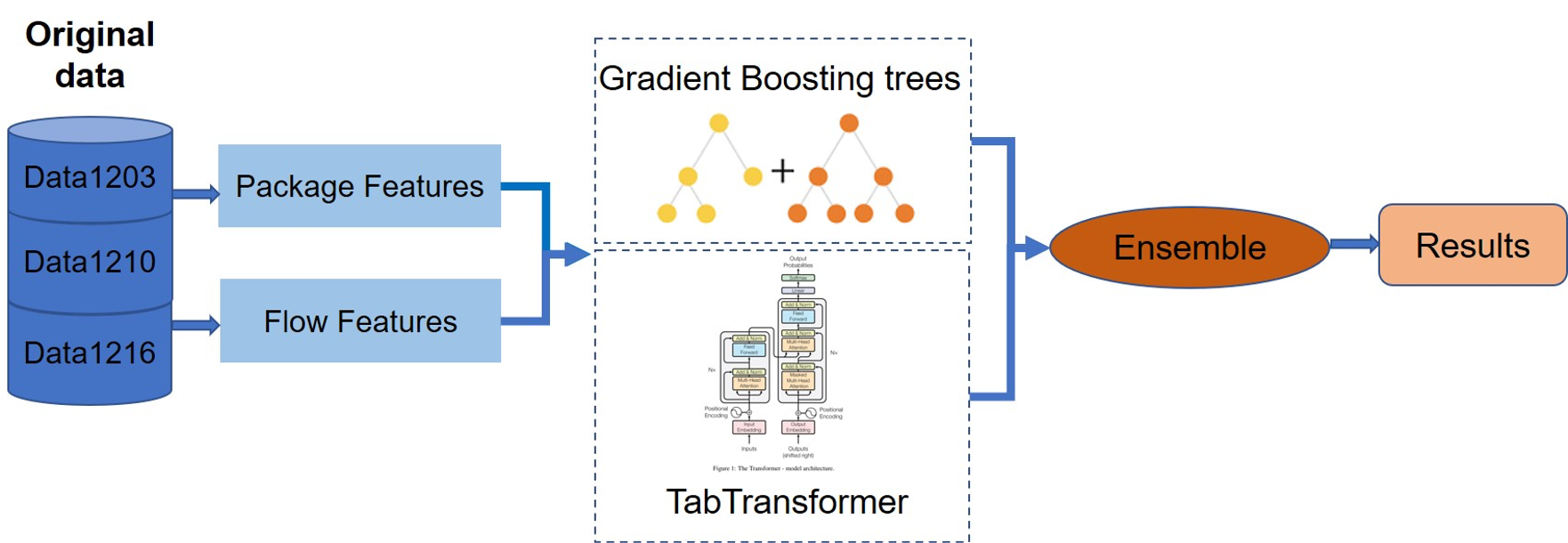
Xinyue Xu, Xiaolu Zheng ICASSP, 2021 Paper In this paper, we present our solution for the ICASSP 2021 Network Anomaly Detection Challenge (NAD) challenge. Our approach ranked as 2nd place in the final leaderboard. |
|
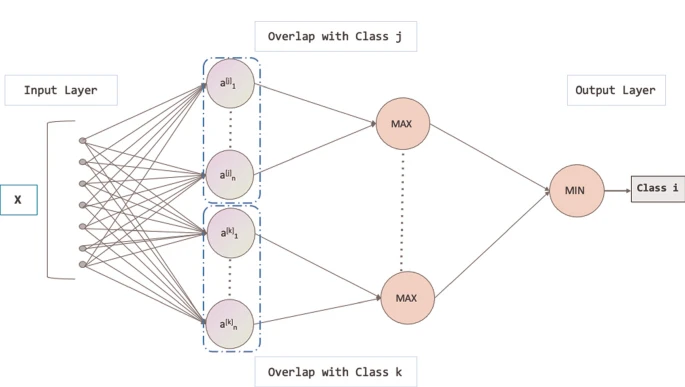
Xinyue Xu, Xiang Ding, Zhenyue Qin, Yang Liu ICONIP, 2021 Paper Interpretability AI4Science We use a variety of classification models to recognize the positive cases of SARS. We conduct evaluation with two types of SARS datasets, numerical and categorical types. For the sake of more clear interpretability, we also generate explanatory rules for the models. |
|
|
|
The source code is from Jon Barron's public academic website. |